Dataset resource
resource.RmdThis resource was established by analyzing several reference ATAC-seq dataset. It includes analysis reports that were generated by ChrAccR. These reports can be used to familiarize oneself with the output of ChrAccR and to explore the analyzed datasets.
ATAC-seq atlas of sorted immune cell populations
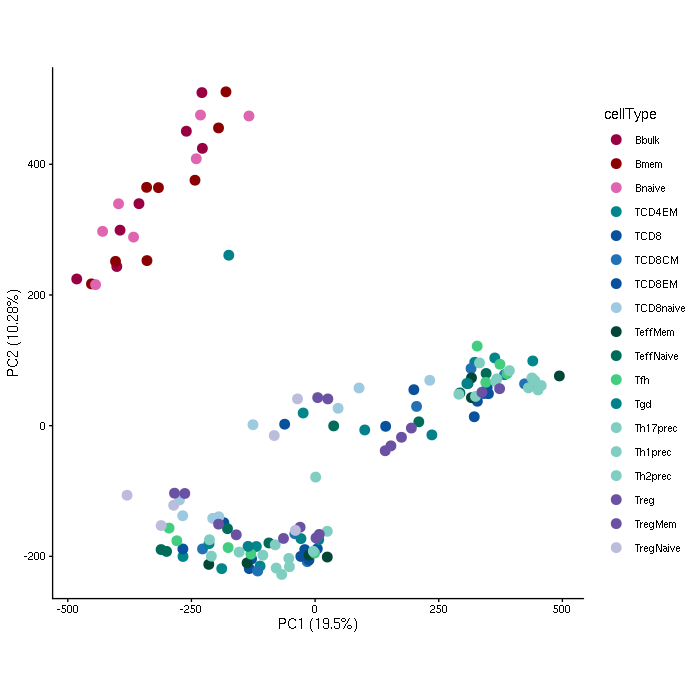
Analysis of sorted immune cells from the B and T cell lineages. The dataset includes ATAC-seq profiles of 142 samples in a naive or an immune-stimulated state. A study providing the dataset and an in-depth analysis has been previously published in Nature Genetics (Calderon et al. 2019).
Human hematopoiesis at single-cell resolution
Analysis of normal human hematopoiesis at single-cell resolution. The dataset comprises 31,894 cells derived from bone marrow and PBMC samples. A study providing the dataset and an in-depth analysis in the context of childhood leukemia has been previously published in Nature Biotechnology (Granja et al. 2019).
